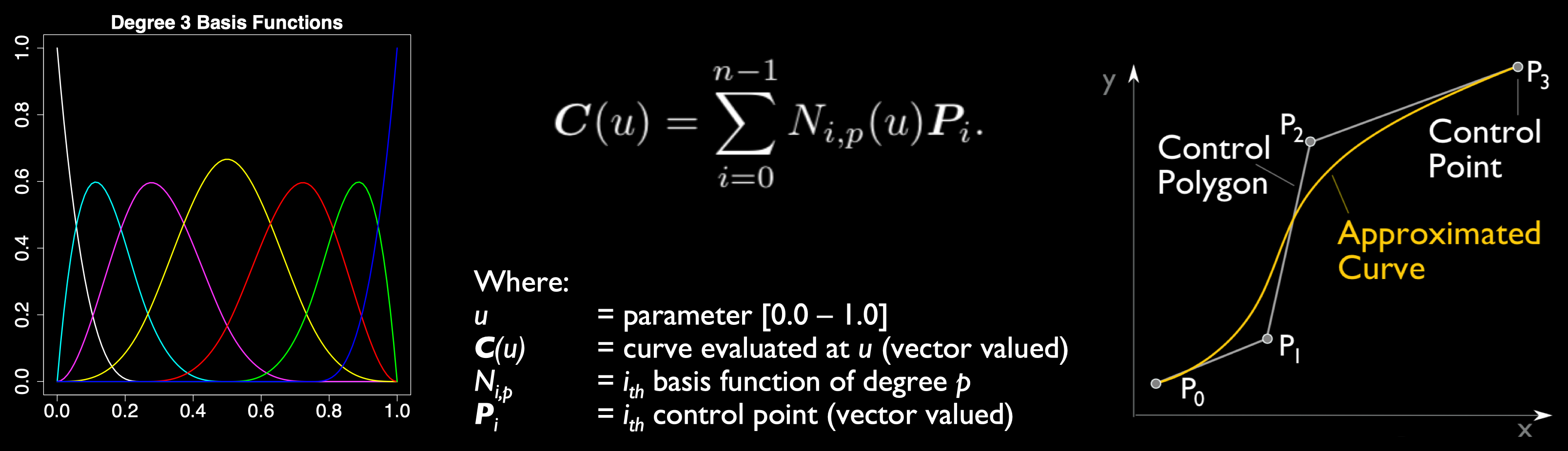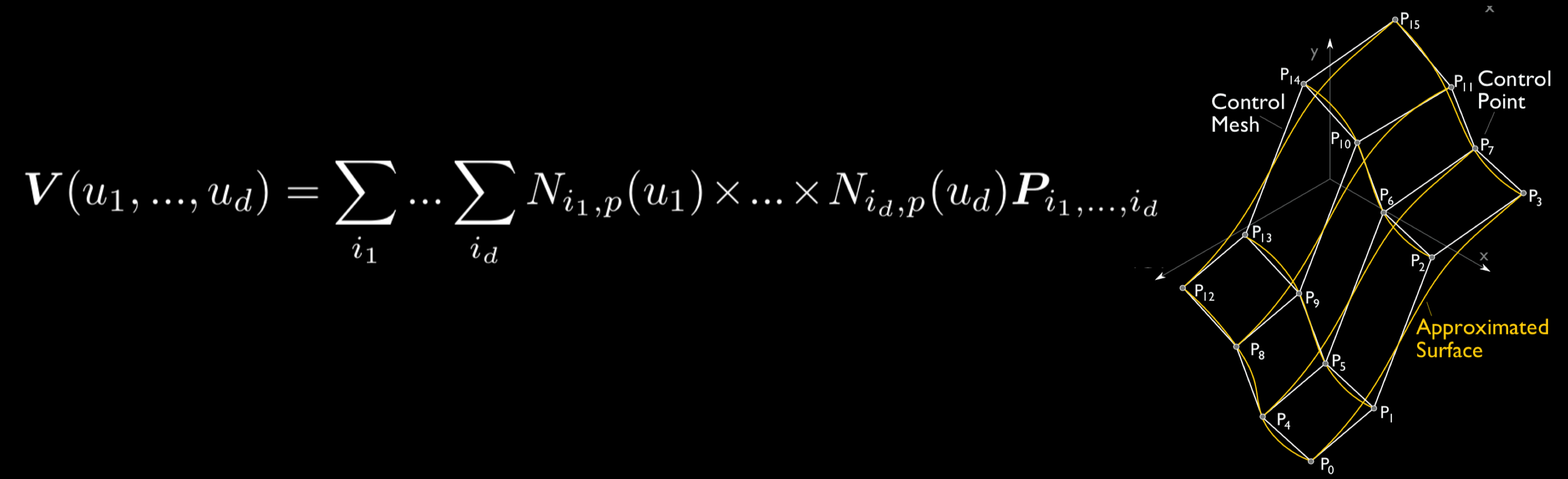
An open-source package of scalable building blocks for data movement tailored to the needs of large-scale parallel analysis workloads
Installation (Linux, Mac, supercomputers, computing clusters):
Download DIY with the following command:
git clone https://github.com/diatomic/diy
and follow the instructions in the README.
Documentation can be found here.
Description:
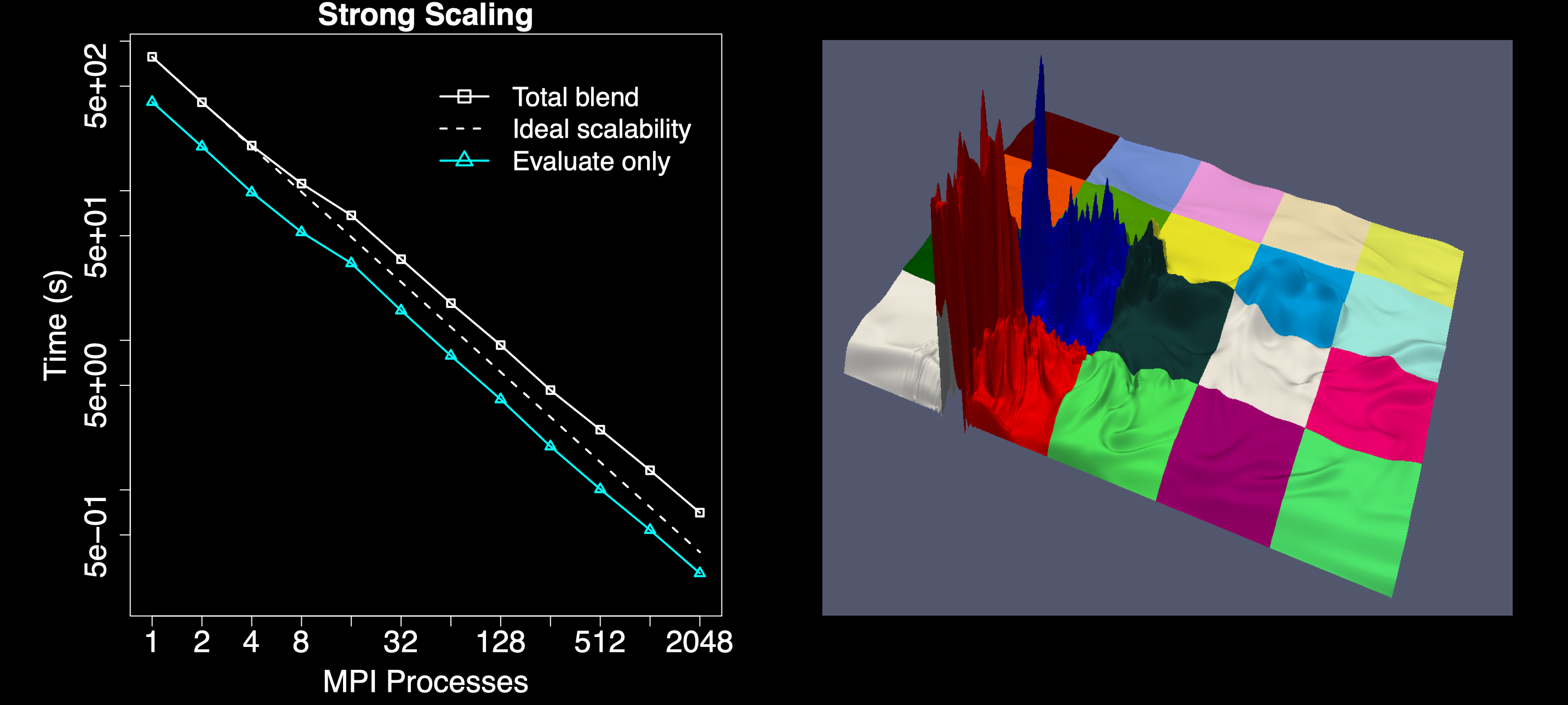
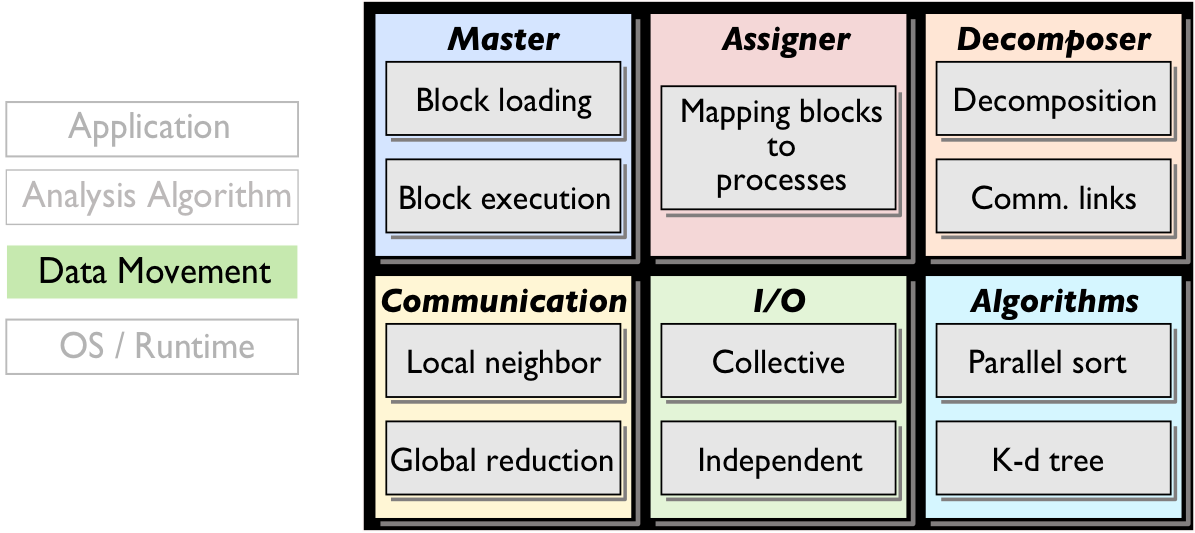
Scalable, parallel analysis of data-intensive computational science relies on the decomposition
of the analysis problem among a large number of data-parallel subproblems, the efficient data
exchange among them, and data transport between them and the memory/storage hierarchy. The
abstraction enabling these capabilities is block-based parallelism; blocks and their message
queues are mapped onto processing elements (MPI processes or threads) and are migrated between
memory and storage by the DIY runtime. Configurable data partitioning, scalable data exchange,
and efficient parallel I/O are the main components of DIY. The current version of DIY has been
completely rewritten to support distributed- and shared-memory parallel algorithms that can run
both in- and out-of-core with the same code. The same program can be executed with one or more
threads per MPI process and with one or more data blocks resident in main memory.
Computational scientists, data analysis researchers, and visualization tool builders can all
benefit from these tools.

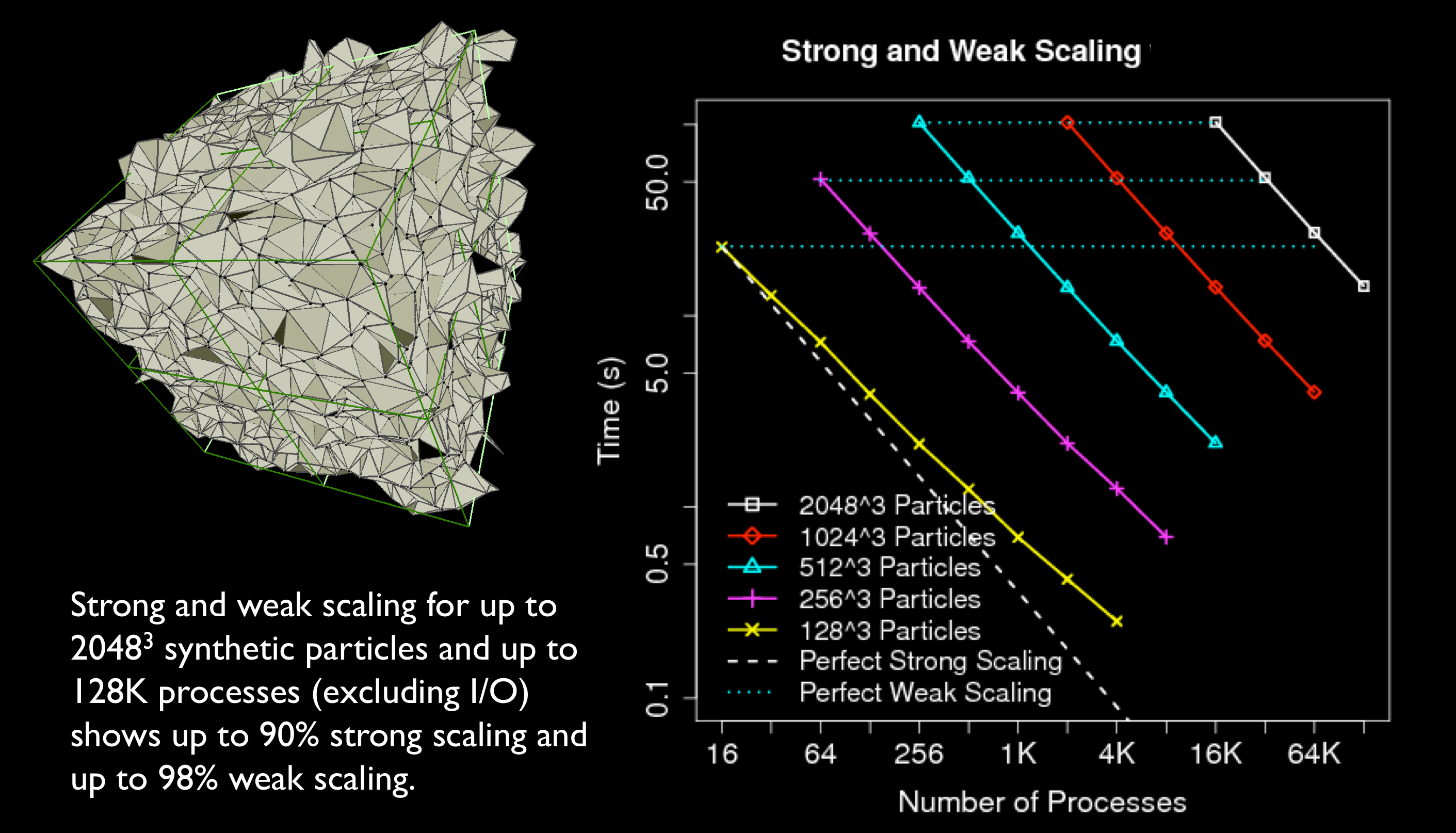
An open-source package for parallelizing Delaunay and Voronoi tessellation over
distributed-memory HPC architecture
Installation (Linux, Mac, supercomputers, computing clusters):
Download Tess with the following command:
git clone https://github.com/diatomic/tess2
and follow the instructions in the README.
Description:
Computing a Voronoi or Delaunay tessellation from a set of points is a core part
of the analysis of many simulated and measured datasets: N-body simulations,
molecular dynamics codes, and LIDAR point clouds are just a few examples. Such
computational geometry methods are common in data analysis and visualization;
but as the scale of simulations and observations surpasses billions of
particles, the existing serial and shared-memory algorithms no longer suffice.
A distributed-memory scalable parallel algorithm is the only feasible approach.
Tess is a parallel Delaunay and Voronoi
tessellation algorithm that automatically determines which neighbor points need
to be exchanged among the subdomains of a spatial decomposition.
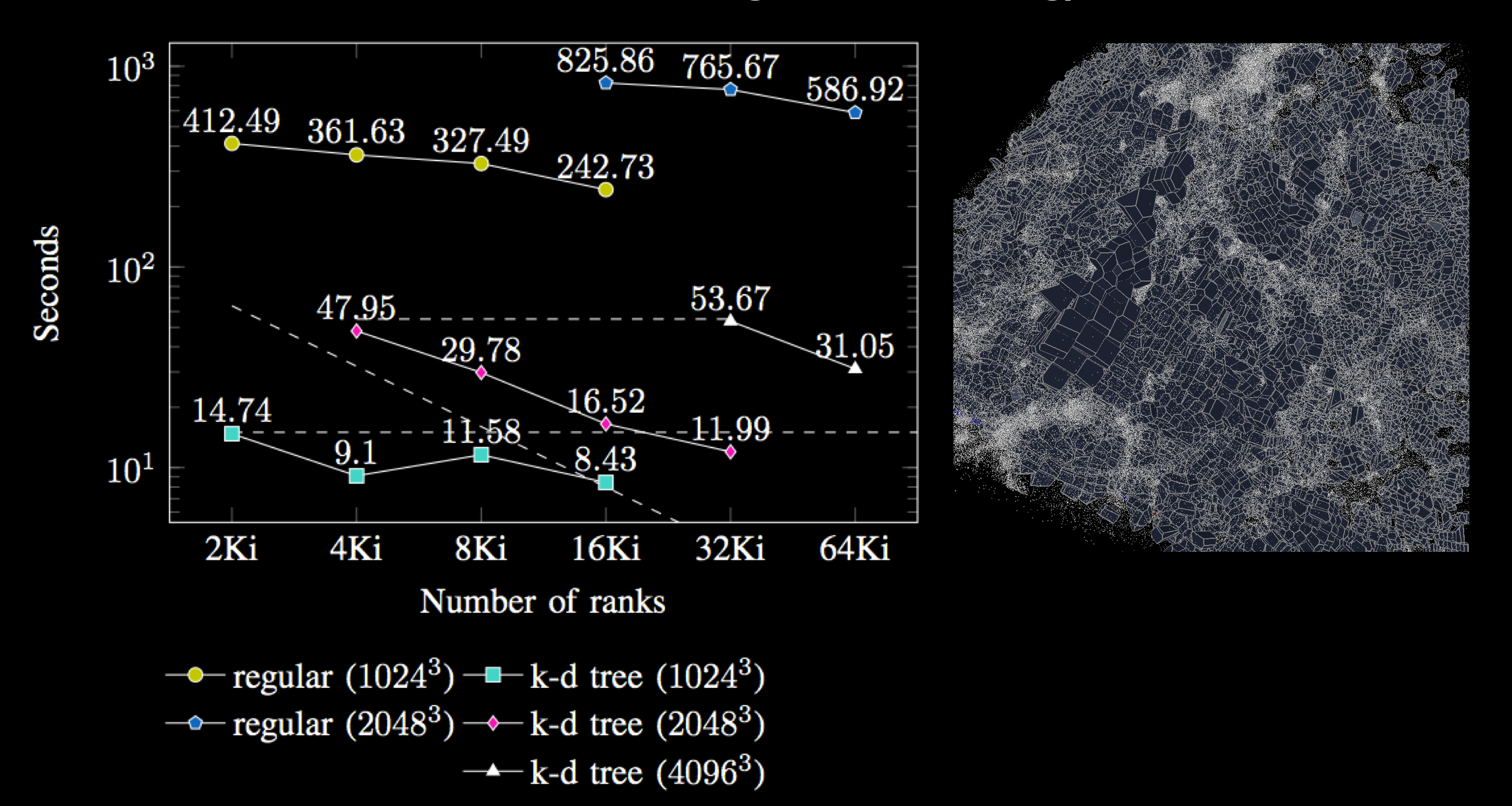
Computing tessellations at scale performs poorly when
the input data is unbalanced, which is why Tess uses k-d trees to evenly
distribute points among processes. The
running times are up to 100 times faster using k-d tree compared with regular
grid decomposition. Moreover, in unbalanced data sets, decomposing the
domain into a k-d tree is up to five times faster than decomposing it into a
regular grid.
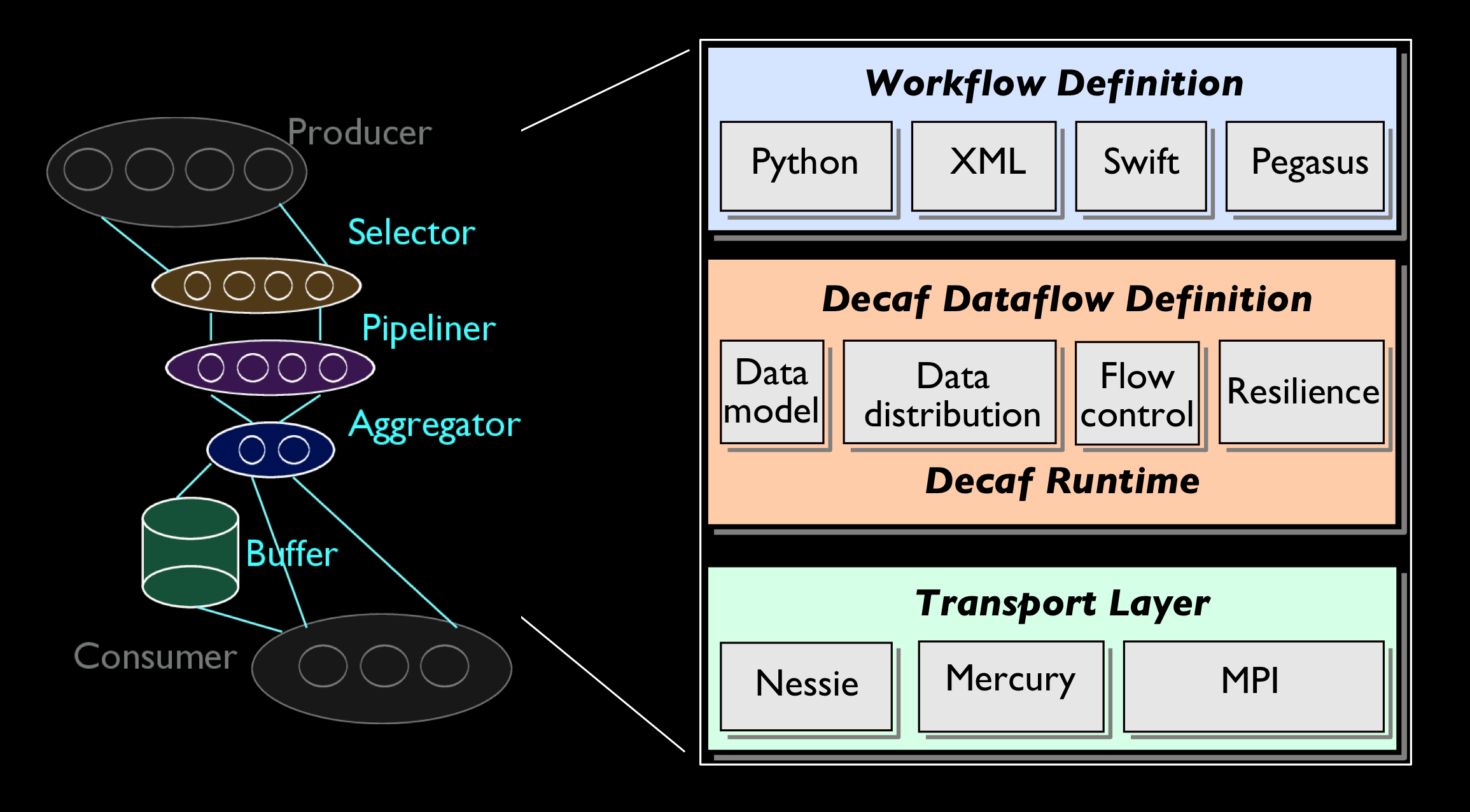
An open-source package of dataflow communication for in situ HPC data analysis
workflows
Installation (Linux, Mac, supercomputers, computing clusters):
Download Decaf with the following command:
git clone https://github.com/tpeterka/decaf
and follow the instructions in the README.
Description:
Decaf is a dataflow system for the parallel communication of coupled tasks in
an HPC workflow. The dataflow can perform arbitrary data transformations ranging
from simply forwarding data to complex data redistribution. Decaf does this by
allowing the user to allocate resources and execute custom code in the dataflow.
All communication through the dataflow is efficient parallel message passing
over MPI. The runtime for calling tasks is entirely message-driven; Decaf
executes a task when all messages for the task have been received. Such a
message-driven runtime allows cyclic task dependencies in the workflow graph,
for example, to enact computational steering based on the result of downstream
tasks. Decaf includes a simple Python API for describing the workflow graph.
This allows Decaf to stand alone as a complete workflow system, but Decaf
can also be used as the dataflow layer by one or more other workflow systems
to form a heterogeneous task-based computing environment.
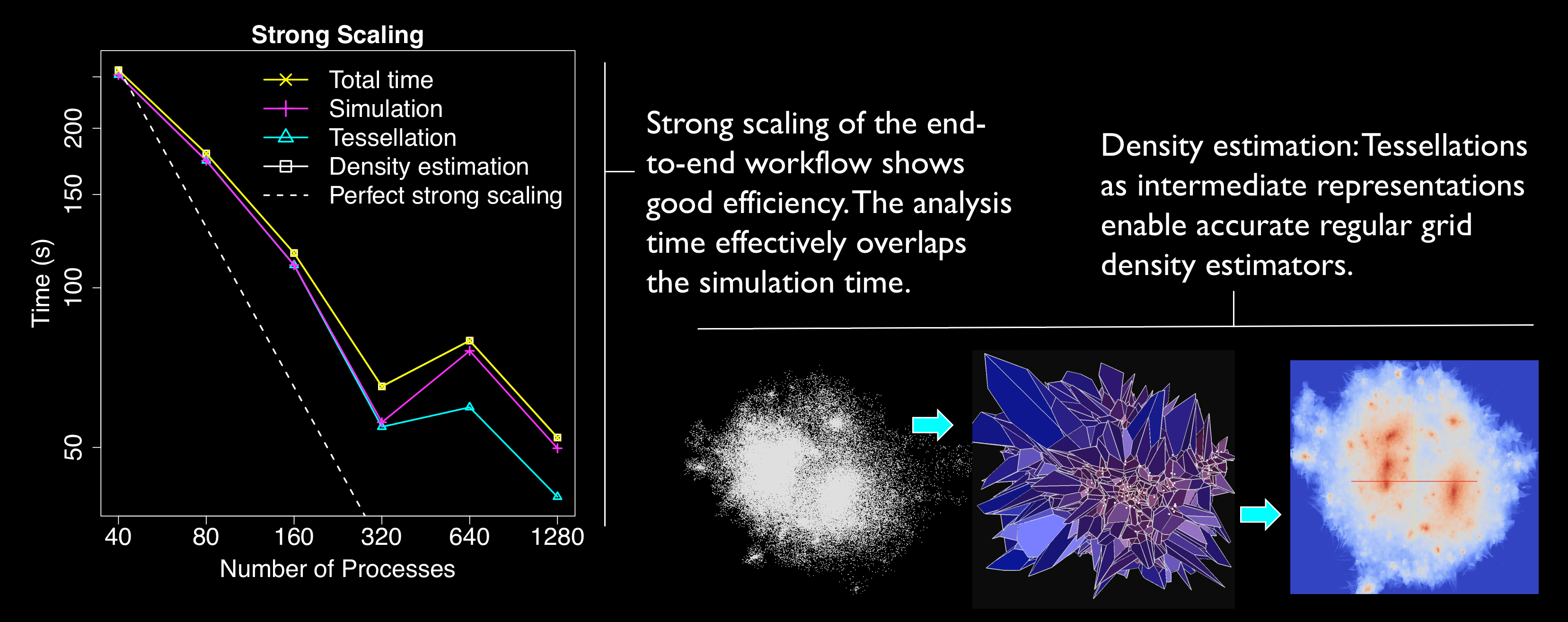
An open-source package of high-performance data transport for in situ HPC workflows
Installation (Linux, Mac, supercomputers, computing clusters):
Download LowFive with the following command:
git clone https://github.com/diatomic/LowFive
and follow the instructions in the README.
Description:
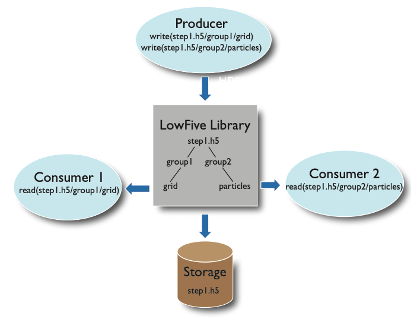
LowFive is a data transport layer based on the HDF5 data model, for in situ workflows. Executables using
LowFive can communicate in situ (using in-memory data and MPI message passing), reading and writing traditional HDF5
files to physical storage, and combining the two modes. Minimal and often no source-code modification is needed for
programs that already use HDF5. LowFive maintains deep copies or shallow references of datasets, configurable by the
user. More than one task can produce (write) data, and more than one task can consume (read) data, accommodating fan-in
and fan-out in the workflow task graph. LowFive supports data redistribution from n producer processes to m consumer
processes.
An open-source in situ HPC workflow management system.
Installation (Linux, Mac, supercomputers, computing clusters):
Download Wilkins with the following command:
git clone https://github.com/orcunyildiz/wilkins
and follow the instructions in the README.
Description:
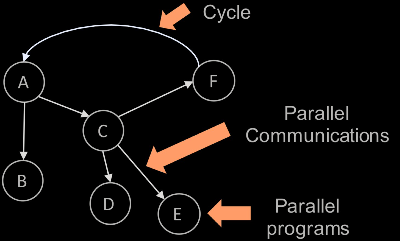
Wilkins is an in situ workflow system that is designed for ease-of-use while providing
scalable and efficient execution of workflow tasks. Wilkins provides a flexible workflow description interface, employs
a high-performance data transport layer based on HDF5, and supports tasks with disparate data rates by providing a
flow control mechanism. Wilkins seamlessly couples scientific tasks that already use HDF5, without requiring task
code modifications.
An open-source package for modeling scientific data with functional approximations based on high-dimensional
multivariate B-spline and NURBS bases
Installation (Linux, Mac, supercomputers, computing clusters):
Download MFA with the following command:
git clone https://github.com/tpeterka/mfa
and follow the instructions in the README.
Description:
Scientific data may be transformed by recasting to a fundamentally different kind of data model than the discrete
point-wise or element-wise datasets produced by computational models. In Multivariate Functional Approximation, or MFA,
scientific datasets are redefined in a hypervolume of piecewise-continuous basis functions. Compared with existing
discrete models, the continuous functional model can save space while affording many of the same spatiotemporal analyses
without reverting back to the discrete form. The MFA model can represent numerous types of data because it is agnostic
to the mesh, field, or discretization of the input dataset. Compared with existing discrete data models, the MFA model
can enable many spatiotemporal analyses, without converting the entire dataset back to the original discrete form. The
MFA often occupies less storage space than the original discrete data, providing some data reduction, depending on data
complexity and intended usage. For example, noise may be intentionally smoothed using a small number of control points
and high-degree basis functions; alternatively, high-frequency data features may be preserved with more control points
and lower degree. Post hoc, the MFA enables analytical closed-form evaluation of points and derivatives, to high order,
anywhere inside the domain, without being limited to the locations of the input data points.